volSegment
Segments volumetric file from a simple threshold which can be set automatically from the otsu estimation.
Usage: volSegment [input] [output]
Allowed options are :
Positionals:
1 TEXT:FILE REQUIRED volumetric input file (.vol, .pgm, .pgm3d, .longvol)
2 TEXT:FILE REQUIRED volumetric output file (.vol, .pgm, .pgm3d, .longvol)
Options:
-h,--help Print this help message and exit.
-i,--input TEXT:FILE REQUIRED volumetric input file (.vol, .pgm, .pgm3d, .longvol)
-o,--output TEXT=result.vol volumetric output file (.vol, .pgm, .pgm3d, .longvol)
--outputTypeUInt to specify the output image type (unsigned int) instead using the default unsigned char. If this flag is selected you have to check the output file format type (longvol, or an ITK image type if the DGtal WITH_ITK option is selected).
--labelBackground option to define a label to regions associated to object background.
-m,--thresholdMin INT=0 min threshold (if not given the max threshold is computed with Otsu algorithm).
-M,--thresholdMax INT=255 max threshold
Example:
You can test the segmentation in the lobster volume file:
$ volSegment ${DGtal}/examples/samples/lobster.vol segmentation.vol -m 70 -M 255
You will obtain a volumetric file representing for each voxel a label associated to a connected component. You can display this segmentation results by extracting it in SDP format with the vol2sdp tool (with option -e to export also the image labels):
$ vol2sdp segmentation.vol segmentation.sdp -e -m 1 -M 255
and display them with 3DSDPViewer :
$ 3dSDPViewer segmentation.sdp --importColorLabels
You should obtain such a result:
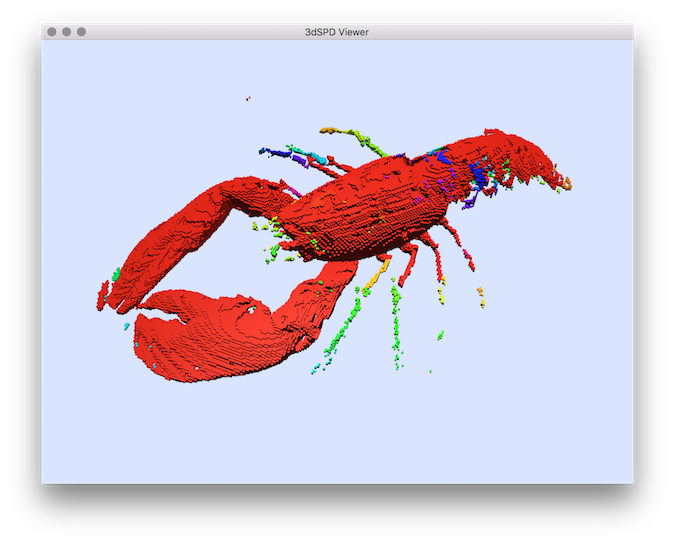
Segmentation result displayed with colors representing the segmentation labels.
- See also
- volSegment.cpp
 1.9.1
1.9.1