Loading...
Searching...
No Matches
Volumetric Tools
3dVolMarchingCubes
Usage: 3dVolMarchingCubes [-i <fileName.vol>] [-t <threshold>] [-a <adjacency>] [-o <output.off>]Outputs the isosurface of value <threshold> of the volume <fileName.vol> as an OFF file <output.off>. The <adjacency> (0/1) allows to choose between interior (6,18) and exterior (18,6) adjacency.Allowed options are :
Example: You should obtain such a result:
 You can test on other samples like https://tc18.org/DataSets/3D_greyscale/bonsai.vol.bz2
You can test on other samples like https://tc18.org/DataSets/3D_greyscale/bonsai.vol.bz2
Positionals:
1 TEXT:FILE REQUIRED the volume file (.vol).
2 TEXT=marching-cubes.off the output OFF file that represents the geometry of the isosurface
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED the volume file (.vol).
-t,--threshold FLOAT=1 the value that defines the isosurface in the image (an integer between 0 and 255).
-a,--adjacency UINT=0 0: interior adjacency, 1: exterior adjacency
-o,--output TEXT=marching-cubes.off the output OFF file that represents the geometry of the isosurface
-n,--noise FLOAT Kanungo noise level in ]0,1[. Note that only the largest connected component is considered and that no specific embedder is used.
$ 3dVolMarchingCubes $DGtal/examples/samples/lobster.vol -t 30
# we invert the default normol orientation to improve display (-n option):
$ meshViewer -i marching-cubes.off -n

Resulting visualization.
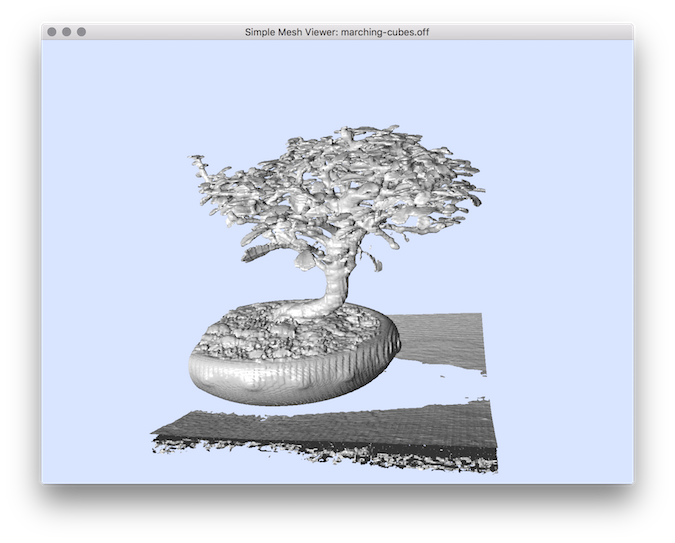
$ 3dVolMarchingCubes -i bonsai.vol -t 80
You should obtain such a result:

Resulting visualization.
- See also
- 3dVolMarchingCubes.cpp
criticalKernelsThinning3D
Usage: criticalKernelsThinning3D [options] –input <3dImageFileName> {vol,longvol,pgm3d...}Allowed options are :
Example:
You should obtain such a result:

Positionals:
1 TEXT:FILE REQUIRED Input vol file.
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input vol file.
-s,--skel TEXT:{ulti,end,isthmus,1isthmus}=1isthmus
Type of skeletonization. Options: 1isthmus, isthmus, end, ulti.
Options: ulti,end, 1isthmus, isthmusulti: delete all voxels except those that change topology.
end: keep voxels with only one neighbor.
1isthmus: keep voxels that are one-isthmus (using LookUpTables) [faster]
isthmus: keep voxels that are one-isthmus or two-isthmus (using LookUpTables) [faster]
-c,--select TEXT:{random,dmax,first}=dmax
Select the ordering for skeletonization. Options: dmax, random, first
Options: dmax, first, random
dmax: Use distance map, selecting voxel with max value.
first: Select first pixel (lexicographical order)
random: Select voxel at random.
-f,--foreground TEXT:{white,black}=black
Foreground color in binary image
-m,--thresholdMin INT=0 Threshold min (excluded) to define binary shape
-M,--thresholdMax INT=255 Threshold max (included) to define binary shape
-p,--persistence INT:POSITIVE=0 Persistence value, implies use of persistence algorithm if p>=1
--profile Profile algorithm
-v,--verbose Verbose output
-o,--exportImage TEXT Export the resulting set of points to a image compatible with GenericWriter.
-e,--exportSDP TEXT Export the resulting set of points in a simple (sequence of discrete point (sdp)).
-t,--visualize Visualize result in viewer
-k,--keepInputDomain Keep the resulting image domain equal to the input image (instead using the resulting bouding box set).
-O,--exportOBJ TEXT Export the resulting set of points in an OBJ file.
-I,--exportInputOBJ TEXT Export the input set of points in an OBJ file.
$ criticalKernelsThinning3D --input ${DGtal}/examples/samples/Al.100.vol --select dmax --skel 1isthmus --persistence 1 -t
Definition ATu0v1.h:57

Resulting visualization.
- See also
- criticalKernelsThinning3D.cpp
homotopicThinning3D
Usage: homotopicThinning3d [options] <3dImageFileName> {vol,longvol,pgm3d...}Allowed options are :
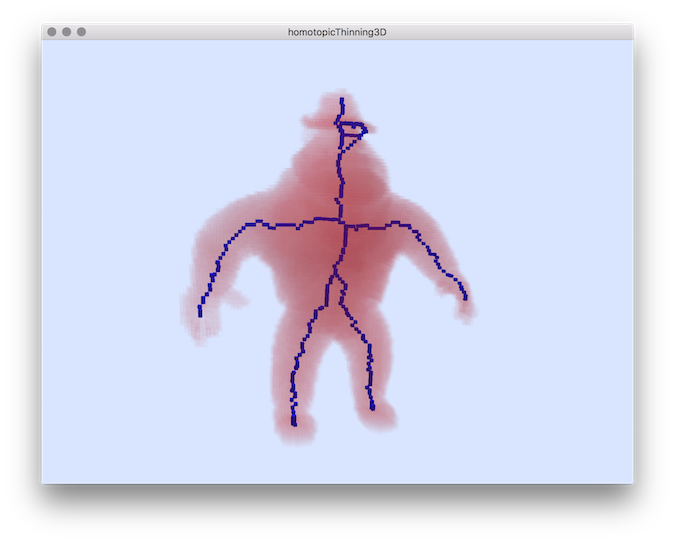
Example: Usage by forcing point to be left by the thinning:You should obtain such a result:
Positionals:
1 TEXT:FILE REQUIRED Input volumetric file (.vol, .pgm3d or p3d)
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input volumetric file (.vol, .pgm3d or p3d)
-m,--min FLOAT=0 Minimum (excluded) value for threshold.
-M,--max FLOAT=255 Maximum (included) value for threshold.
-e,--exportSDP TEXT Export the resulting set of points in a simple (sequence of discrete point (sdp)).
--fixedPoints INT ... defines the coordinates of points which should not be removed.
-s,--fixedPointSDP TEXT:FILE use fixed points from a file.
$ homotopicThinning3D ${DGtal}/examples/samples/Al.100.vol --fixedPoints 56 35 5 56 61 5 57 91 38 58 8 38 45 50 97

Resulting visualization.
- See also
- homotopicThinning3D.cpp
volAddBorder
Usage: volAddBorder <volFileName> <volOutputFileName>Allowed options are :
Example: You should obtain an resulting image with domain: -1, -1, -1 x 100, 100, 100 instead 0, 0, 0 x 99, 99, 99
Positionals:
1 TEXT:FILE REQUIRED Input vol file.
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input vol file.
--inside Sets zero value to domain boundary voxels without changing the domain extent.
-o,--output TEXT Output filename.
$ volAddBorder -i $DGtal/examples/samples/Al.100.vol -o Al.100border.vol
- See also
- volAddBorder.cpp
volAddNoise
Allowed options are:
Example: You should obtain such a visualization:
Positionals:
1 TEXT:FILE REQUIRED input image file name (any 3D image format accepted by DGtal::GenericReader)
2 TEXT=result.vol output image file name (any 3D image format accepted by DGtal::GenericWriter)
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED input image file name (any 3D image format accepted by DGtal::GenericReader)
-o,--output TEXT=result.vol output image file name (any 3D image format accepted by DGtal::GenericWriter)
-n,--noise FLOAT=0.5 Kanungo noise level in ]0,1[ (default 0.5)
-m,--max Extract only the largest 6-connected component.
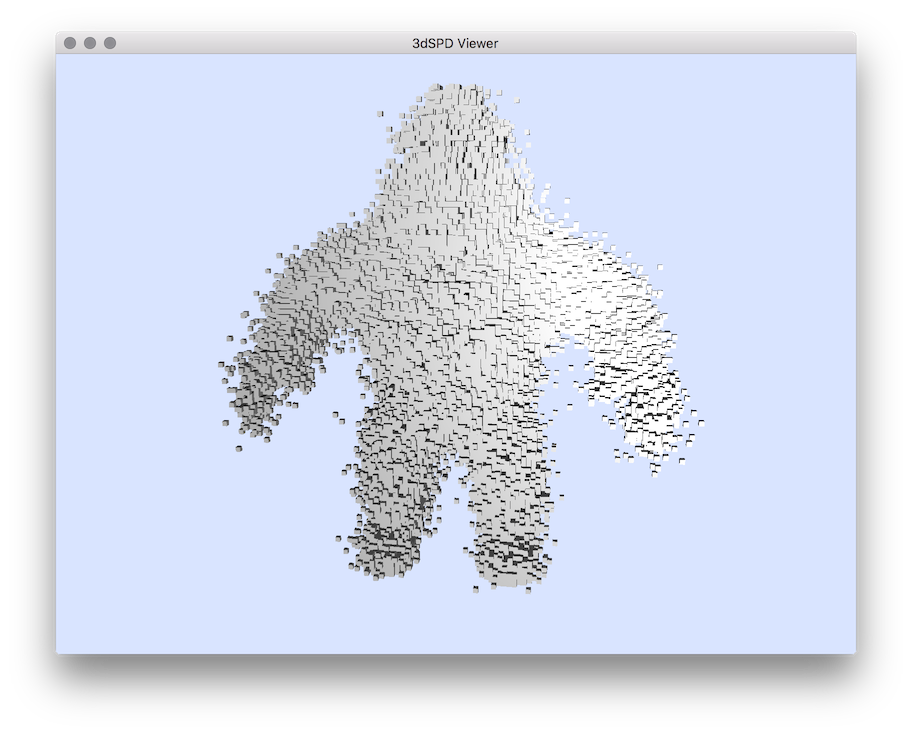
$ volAddNoise $DGtal/examples/samples/Al.100.vol AlNoisy0.4.vol -n 0.4
# Converting in sdp to display:
$ vol2sdp AlNoisy0.4.vol AlNoisy0.4.sdp
# displaying sequence of points:
$ 3dSDPViewer AlNoisy0.4.sdp

Resuling visualisation with 3dSDPViewer.
- See also
- volAddNoise.cpp
volCComponentCounter
Allowed options are :
Example: You should obtain:
Positionals:
1 TEXT:FILE REQUIRED volume file (.vol).
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED volume file (.vol).
-c,--connectivity UINT:{6,18,26}=6 object connectivity (6,18,26) (default: 6 ).
$ volCComponentCounter $DGtal/examples/samples/Al.100.vol
New Block [Initial disjoint sets construction] EndBlock [Initial disjoint sets construction] (515.602 ms) New Block [Merging neighboring sets] EndBlock [Merging neighboring sets] (1672.21 ms) Number of disjoint 6-components = 2
- See also
- volCComponentCounter.cpp
volCrop
Usage: volCrop –input <volFileName> –o <volOutputFileName> (both files can be independently in vol, pgm3D, p3d format)Allowed options are :
Example: You should obtain such a visualization:
Positionals:
1 TEXT:FILE REQUIRED Input vol file.
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input vol file.
-o,--output TEXT=output.vol Output filename.
--xMin INT=0 x coordinate of lower point.
--yMin INT=0 y coordinate of lower point.
--zMin INT=0 z coordinate of lower point.
--xMax INT REQUIRED x coordinate of upper point.
--yMax INT REQUIRED y coordinate of upper point.
--zMax INT REQUIRED z coordinate of upper point.
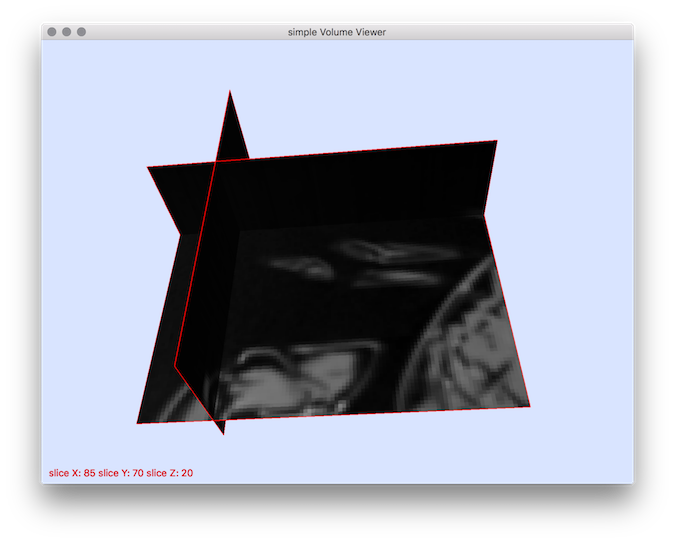
$ volCrop --xMin 50 --yMin 50 --zMin 10 --xMax 150 --yMax 150 --zMax 50 -i ${DGtal}/examples/samples/lobster.vol -o croppedLobster.vol
$ ./visualisation/3dVolViewer croppedLobster.vol -m 60

Resulting visualization.
- See also
- volCrop.cpp
volFillInterior
Allowed options are :
Example:
Positionals:
1 TEXT:FILE REQUIRED Input vol file.
2 TEXT=result.vol Output filename.
3 UINT Set the filling value other than the default value of 128.
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input vol file.
-o,--output TEXT=result.vol Output filename.
-v,--fillValue UINT Set the filling value other than the default value of 128.
$ volFillInterior -i ${DGtal}/examples/samples/lobster.vol -o filled.vol
- See also
- volFillInterior
volFlip
Allowed options are :
Example: You should obtain such a result:

Positionals:
1 TEXT:FILE REQUIRED Input vol filename.
2 TEXT:FILE REQUIRED Output vol filename.
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input vol filename.
--imagePlane UINT=[0,1] x 2 arg= {0,1,2} x {0,1,2} defines the axis of the slice image which will be transformed (by default arg= 0 1 i.e. the slice image defined in the X,Y plane (Z=cst)
--flipDimension UINT=0 specify which axis will be used to apply the flip.
-o,--output TEXT=result.vol Output filename.
$ volFlip --imagePlane 0 1 --flipDimension 0 -i ${DGtal}/examples/samples/lobster.vol -o flippedXxyLobster.vol

(a) source image (b) flipped version.
- See also
- volFlip.cpp
volImageMetrics
Allowed options are :
Example: You should obtain such an output:
Positionals:
1 TEXT:FILE REQUIRED Input filename of volume A (vol format, and other pgm3d can also be used).
2 TEXT:FILE REQUIRED Input filename of volume B (vol format, and other pgm3d can also be used).
Options:
-h,--help Print this help message and exit
-a,--volA TEXT:FILE REQUIRED Input filename of volume A (vol format, and other pgm3d can also be used).
-b,--volB TEXT:FILE REQUIRED Input filename of volume B (vol format, and other pgm3d can also be used).
# generating another input vol file using tutorial example (eroded.vol):
$DGtal/build/examples/tutorial-examples/FMMErosion
# compare the two images:
$ volImageMetrics eroded.vol $DGtal/examples/samples/cat10.vol
# Image based measures (generated with volImageMetrics) given with the image A: eroded.voland the image B: /Users/kerautre/EnCours/DGtal/examples/samples/cat10.vol # RMSE PSNR 33.9411 171.331
- See also
- volImageMetrics.cpp
volInfo
Usage: ./volumetric/volInfo [OPTIONS] 1 [2]Allowed options are :
Example: You can retrieve information from the vol file as:
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input vol file.
Positionals:
1 TEXT:FILE REQUIRED Input vol file.
$ volInfo $DGtal/examples/samples/lobster.vol
- See also
- volInfo.cpp
volIntensityScale
Allowed options are :
Example: In this example, we select all intensities included in [0,100] and scale them into [0, 255]:You should obtain such a result:

Positionals:
1 TEXT:FILE REQUIRED Input vol file.
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input vol file.
-o,--output TEXT=result.vol volumetric output file (.vol, .pgm, .pgm3d, .longvol)
-m,--inMin INT=0 the min value of the input image.
-M,--inMax INT=255 the max value of the input image.
--outMin INT=0 the min value of the output image.
--outMax INT=255 the max value of the output image.
$ volIntensityScale ${DGtal}/examples/samples/lobster.vol --inMin 0 --inMax 100 lobster0-100.vol

(a) source image, (b) intensity scaled image.
- See also
- volIntensityScale.cpp
volMask
Usage: volMask [input]Allowed options are :
Example:
Positionals:
1 TEXT:FILE REQUIRED an input 3D image vol (or ITK: .nii, mha, ... ) file.
2 TEXT=result.vol the output masked image.
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED an input 3D image vol (or ITK: .nii, mha, ... ) file.
-t,--inputType TEXT to specify the input image type (int or double).
-a,--mask TEXT:FILE the mask image that represents the elements that are copied as output in the resulting image (by default set to 1 you can change this value by using --maskValue).
-o,--output TEXT=result.vol the output masked image.
-f,--offsetBorder UINT=0 add a border offset to the bounding box of the masked value domain.
-m,--maskValue INT=1 the masking value.

Example of result.
- See also
- volMask.cpp
volReSample
Allowed options are :
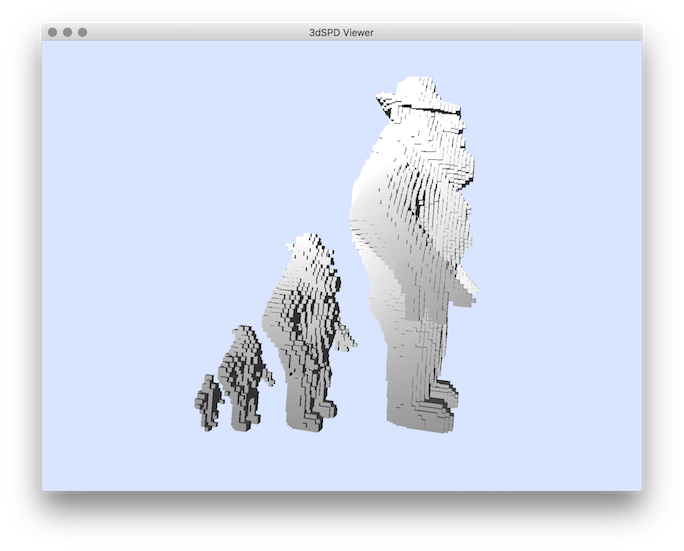
Example: Here is an example of re sampling with different grid sizes 2, 4 and 8:We can convert the resulting volumetric files into a sequence of discrete points with the tool vol2sdp : Note that if DGtal is compiled with the option DGTAL_WITH_ITK set to ON, you can export the image in format ITK format and integrating image spacing.You should obtain such a result:
Positionals:
1 TEXT:FILE REQUIRED input volumetric file (.vol, .longvol, .pgm3d).
2 TEXT the new volumetric file (.vol, .longvol, .pgm3d).
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED input volumetric file (.vol, .longvol, .pgm3d).
-o,--output TEXT the new volumetric file (.vol, .longvol, .pgm3d).
-g,--gridSize FLOAT x 3 size_x size_y size_z : the grid size of the re sampling
$ volReSample $DGtal/examples/samples/Al.100.vol AlRS2.vol -g 2 2 2
$ volReSample $DGtal/examples/samples/Al.100.vol AlRS4.vol -g 4 4 4
$ volReSample $DGtal/examples/samples/Al.100.vol AlRS8.vol -g 8 8 8
$ vol2sdp $DGtal/examples/samples/Al.100.vol -m 1 -o AlRS1.sdp
$ vol2sdp AlRS2.vol -m 1 -o AlRS2.sdp
$ vol2sdp AlRS4.vol -m 1 -o AlRS4.sdp
$ vol2sdp AlRS8.vol -m 1 -o AlRS8.sdp
$ cat AlRS{1,2,4,8}.sdp >> AlRS1_2_4_8.sdp
# display the resulting file:
3dSDPViewer AlRS1_2_4_8.sdp

Resulting of re sampling with grid size = 2, 4 and 8.
- See also
- volReSample.cpp
volSegment
Usage: volSegment [input] [output]Allowed options are :
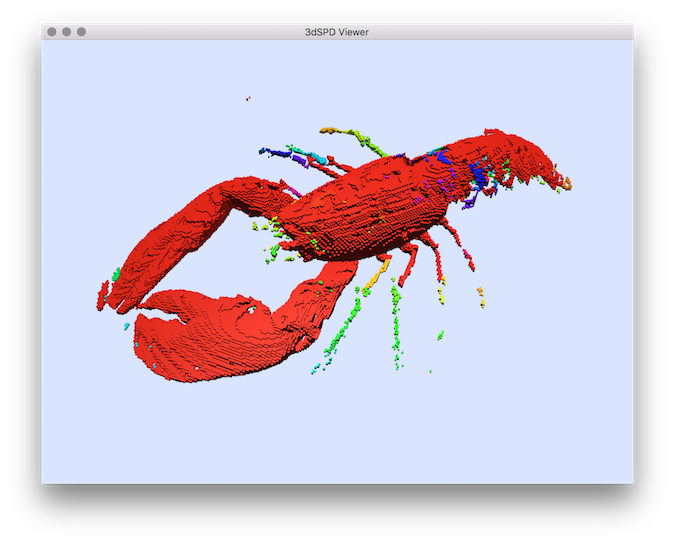
Example: You can test the segmentation in the lobster volume file:You will obtain a volumetric file representing for each voxel a label associated to a connected component. You can display this segmentation results by extracting it in SDP format with the vol2sdp tool (with option -e to export also the image labels): and display them with Doc3DSDPViewer :You should obtain such a result:
Positionals:
1 TEXT:FILE REQUIRED volumetric input file (.vol, .pgm, .pgm3d, .longvol)
2 TEXT:FILE REQUIRED volumetric output file (.vol, .pgm, .pgm3d, .longvol)
Options:
-h,--help Print this help message and exit.
-i,--input TEXT:FILE REQUIRED volumetric input file (.vol, .pgm, .pgm3d, .longvol)
-o,--output TEXT=result.vol volumetric output file (.vol, .pgm, .pgm3d, .longvol)
--outputTypeUInt to specify the output image type (unsigned int) instead using the default unsigned char. If this flag is selected you have to check the output file format type (longvol, or an ITK image type if the DGtal DGTAL_WITH_ITK option is selected).
--labelBackground option to define a label to regions associated to object background.
-m,--thresholdMin INT=0 min threshold (if not given the max threshold is computed with Otsu algorithm).
-M,--thresholdMax INT=255 max threshold
$ volSegment ${DGtal}/examples/samples/lobster.vol segmentation.vol -m 70 -M 255
$ vol2sdp segmentation.vol segmentation.sdp -e -m 1 -M 255
$ 3dSDPViewer segmentation.sdp --importColorLabels

Segmentation result displayed with colors representing the segmentation labels.
- See also
- volSegment.cpp
volShapeMetrics
Usefull to determine classical statistics like false positive related stats.Usage: volShapeMetrics –volA <volAFilename> –volB <volBFilename>Allowed options are :
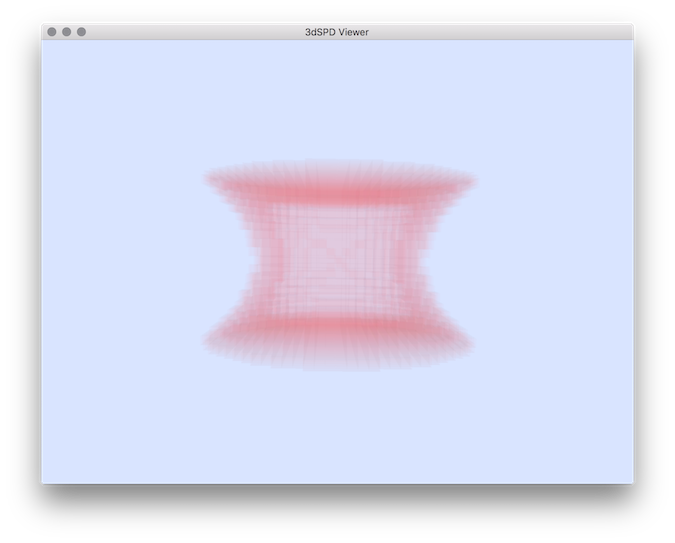
Example: To test this tool, we need to generate a volumetric file to be compared to an original one: Then we can apply comparisons of the two shapes:You should obtain different comparison measures and you can display the set of voxels associated to the false positive ( falsePos.sdp ):
Positionals:
1 TEXT:FILE REQUIRED Input filename of volume A (vol format, and other pgm3d can also be used).
2 TEXT:FILE REQUIRED Input filename of volume B (vol format, and other pgm3d can also be used).
Options:
-h,--help Print this help message and exit
-a,--volA TEXT:FILE REQUIRED Input filename of volume A (vol format, and other pgm3d can also be used).
-b,--volB TEXT:FILE REQUIRED Input filename of volume B (vol format, and other pgm3d can also be used).
--aMin INT=0 min threshold for a voxel to be considered as belonging to the object of volume A. (default 0)
--aMax INT=128 max threshold for a voxel to be considered as belonging to the object of volume A. (default 128)
--bMin INT=0 min threshold for a voxel to be considered as belonging to the object of volume B. (default 0)
--bMax INT=128 max threshold for a voxel to be considered as belonging to the object of volume B. (default 128)
--noDistanceComparisons to avoid to apply distance map computation if the distance comparaison are not needed.
--distancesFromBnotInAOnly apply distance map measures only for voxels of B which are not in A (else the measure are given from all distances of the object B).
--displayTFstats Change the comparison diplay by using the true/false/positive/negative notation and considering the shape A as reference. It also display precision/recall/f-mean statistics.
--exportSDP Export voxels belonging to each categorie (voxels of ( B in A) , (NOT in B and NOT in A), (B and NOT in A) and (Voxels of NOT in B and in A)).
# generation of the file "eroded.vol" from DGtal examples:
$ $DGtal/build/examples/tutorial-examples/FMMErosion
$ volShapeMetrics -a eroded.vol --aMin 1 --aMax 255 -b $DGtal/examples/samples/cat10.vol --bMin 1 --bMax 255 --displayTFstats --exportSDP
$ 3dSDPViewer -i falsePos.sdp -c 250 40 40 5

Resulting False positive set of voxels.
- See also
- volShapeMetrics.cpp
volSubSample
Usage: ./volumetric/volSubSample [OPTIONS] 1 [2]Allowed options are :
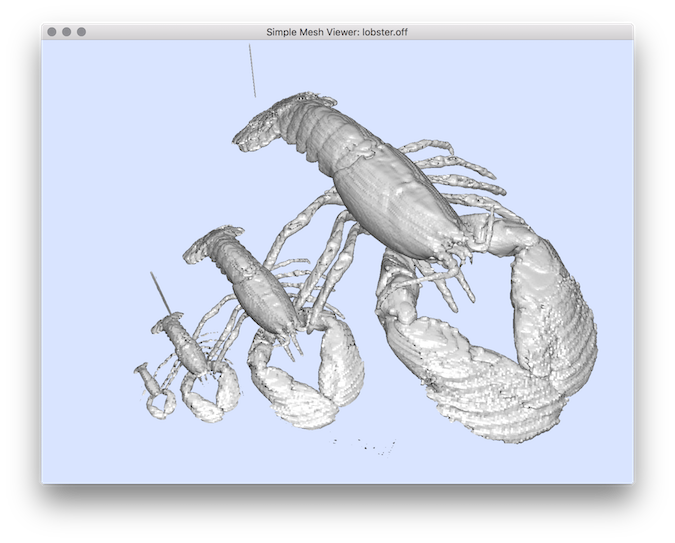
Example: You can apply several sub sampling: You can display the result by extracting the surface using 3dVolMarchingCubes:You should obtain such a result:
Positionals:
1 TEXT:FILE REQUIRED Input vol file.
2 TEXT=result.vol Output filename.
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input vol file.
-o,--output TEXT=result.vol Output filename.
-f,--function TEXT:{mean,none,max,min,mean}=mean
Function used to the down-sampling: {none,max, min, mean}
Positionals:
1 TEXT:FILE REQUIRED Input vol file.
2 TEXT=result.vol Output filename.
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input vol file.
-o,--output TEXT=result.vol Output filename.
-f,--function TEXT:{mean,none,max,min,mean}=mean
Function used to the down-sampling: {none,max, min, mean}
$ volSubSample $DGtal/examples/samples/lobster.vol lobster2.vol -f mean
$ volSubSample lobster2.vol lobster4.vol -f mean
$ volSubSample lobster4.vol lobster8.vol -f mean
$ 3dVolMarchingCubes $DGtal/examples/samples/lobster.vol lobster.off -t 30
$ 3dVolMarchingCubes lobster2.vol -t 30 lobster2.off
$ 3dVolMarchingCubes lobster4.vol -t 30 lobster4.off
$ 3dVolMarchingCubes lobster8.vol -t 30 lobster8.off
$ meshViewer lobster.off lobster2.off lobster4.off lobster8.off -n

Resulting visualization.
- See also
- volSubSample.cpp
volTrValues
Usage: volTrValues –input <volFileName> –o <volOutputFileName> -s 1 99 -r 100 200
=> all voxels of values 1 (resp. 99) will be 100 (resp. 200) in the resulting image.
Allowed options are :
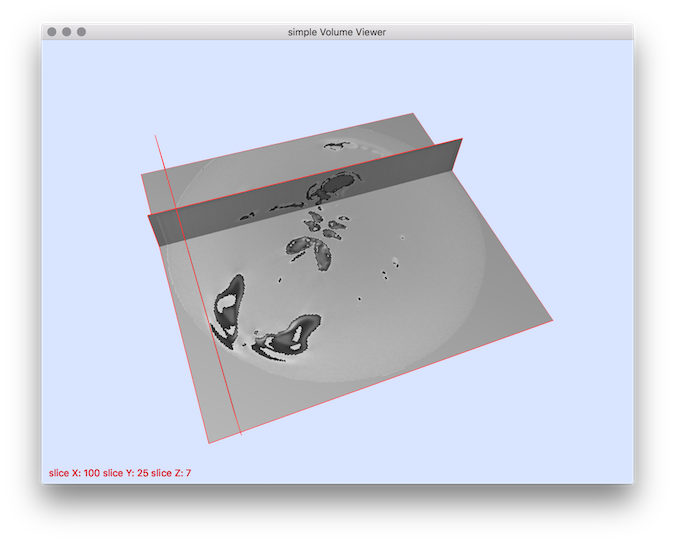
Example: This tool can be useful to apply simple intensity transforms. For instance if you want to transform all intensities starting from 0 to 50 into interval 200 250 you can do as follows:By using 3dImageViewer ou should obtain such a result:
=> all voxels of values 1 (resp. 99) will be 100 (resp. 200) in the resulting image.
Allowed options are :
Positionals:
1 TEXT:FILE REQUIRED Input vol file.
Options:
Positionals:
1 TEXT:FILE REQUIRED Input vol file.
2 TEXT=result.vol Output filename.
Options:
-h,--help Print this help message and exit
-i,--input TEXT:FILE REQUIRED Input vol file.
-o,--output TEXT=result.vol Output filename.
-s,--inputVals UINT ... REQUIRED specify the values which will be transformed with the output values (given with --outputVals).
-r,--outputVals UINT ... REQUIRED specify the values which will be transformed with the output values (given with --outputVals).
$ volTrValues $DGtal/examples/samples/lobster.vol lobsterTr.vol -s {0..50} -r {200..250}
$ 3dImageViewer lobsterTr.vol

Result visualization.
- See also
- volTrValues.cpp